AbbVie is committed to advancing research in NSCLC
EXPLORE MET TESTING METHODOLOGIES IN NSCLC
*Estimated prevalence in non-squamous NSCLC. Imagery is for illustrative purposes only and does not
proportionally represent biomarker prevalence as shown.
MET Biomarker Testing Methodologies
IHC is an established methodology used to detect protein biomarkers such as PD-L1.3,7,9 NGS and FISH are examples of genetic testing modalities that can detect MET amplification and/or METex14 skipping.5,7
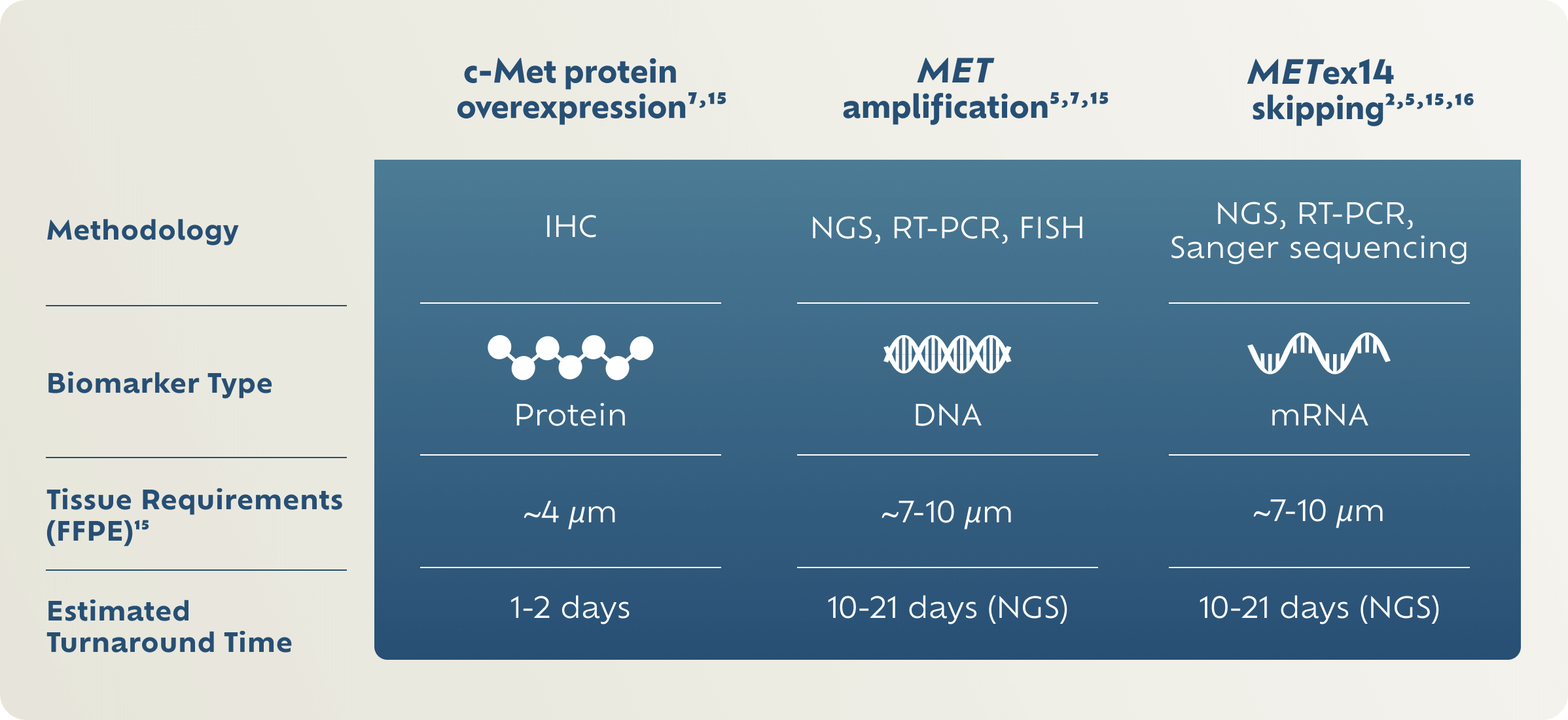
There are no FDA-approved tests for c-Met protein overexpression or MET gene amplification.
FFPE, formalin-fixed paraffin-embedded; FISH, fluorescence in situ hybridization; IHC, immunohistochemistry; NGS, next-generation sequencing; RT-PCR, reverse transcription-polymerase chain reaction.
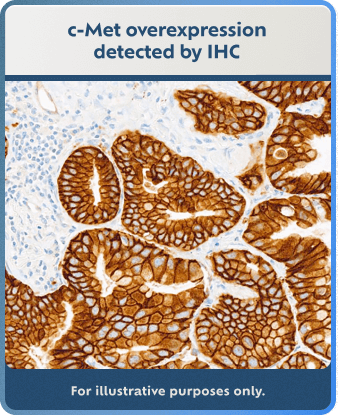
Explore MET Detection Methods
IHC, NGS, and FISH detection method examples are for illustrative purposes only.
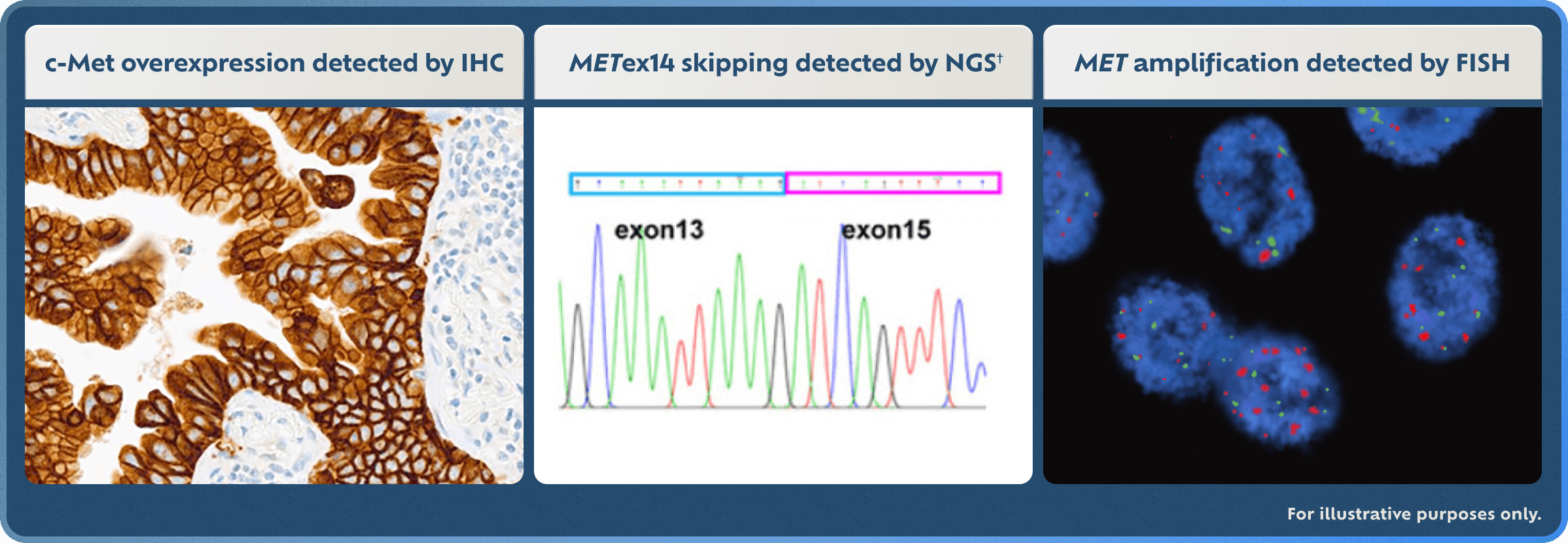
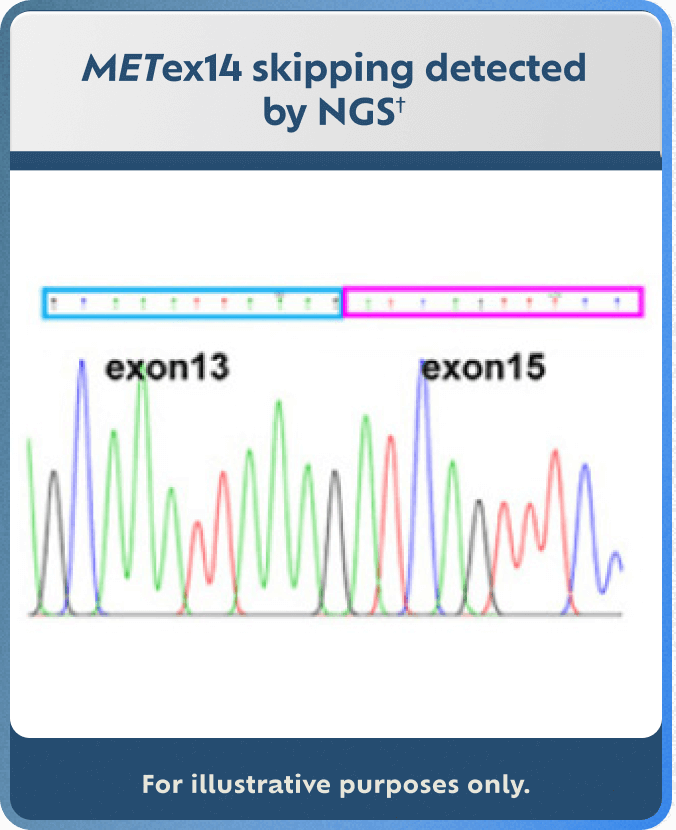
†Adapted with permission from M Shi, et al. Cancer Genetics. 2021;256-257:62-67.17

c-Met protein overexpression and MET gene amplification are emerging biomarkers in clinical research as potential therapeutic targets.
There are no FDA-approved tests for c-Met protein overexpression or MET gene amplification.
Biomarkers included are select MET aberrations that are both emerging and established biomarkers in NSCLC. This is not an exhaustive list of all NSCLC biomarkers and selected MET aberrations may overlap with other NSCLC biomarkers.
MET aberration prevalence estimates and prognosis statements are based on multiple sources. Survival and prevalence data can vary among studies and data sets because of detection methodology used, patient sample sizes, and/or demographic characteristics. Some patients may have more than one MET aberration.
REFERENCES: 1. Motwani M, Panchabhai S, Bar J, et al. Prevalence of c-Met overexpression (c-Met+) and impact of prior lines of treatment on c-Met protein expression in NSCLC. J Thorac Oncol. 2021;16(10):S1169-S1170. 2. Liang H, Wang M. MET oncogene in non-small cell lung cancer: mechanism of MET dysregulation and agents targeting the HGF/c-Met axis. Onco Targets Ther. 2020;13:2491-2510. 3. Zhang Y, Du Z, Zhang M. Biomarker development in MET-targeted therapy. Oncotarget. 2016;7(24):37370-37389. 4. Kerr K, Bibeau F, Thunnissen E, et al. The evolving landscape of biomarker testing for non-small cell lung cancer in Europe. Lung Cancer. 2021;151:161-175. 5. Drilon A, Cappuzzo F, Ou SI, Camidge DR. Targeting MET in lung cancer: will expectations finally be MET? J Thorac Oncol. 2017;12(1):15-26. 6. Tong JH, Yeung SF, Chan AW, et al. MET amplification and exon 14 splice site mutation define unique molecular subgroups of non-small cell lung carcinoma with poor prognosis. Clin Cancer Res. 2016;22(12):3048-3056. 7. Ansell P, Baijal S, Liede A, et al. Prevalence and characterization of c-MET-overexpressing non-small cell lung cancer (NSCLC) across clinical trial samples and real-world patient cohorts from the City of Hope National Medical Center. Poster presented at: Cancer Research UK Lung Cancer Conference; November 15-17, 2022; Manchester, United Kingdom. 8. Organ SL, Tsao MS. An overview of the c-MET signaling pathway. Ther Adv Med Oncol. 2011;3(1 Suppl):S7-S19. 9. Mino-Kenudson M. Immunohistochemistry for predictive biomarkers in non-small cell lung cancer. Transl Lung Cancer Res. 2017;6(5):570-587. 10. Bar J, Cai MH, Choi YC, et al. Prevalence, molecular characterization, and prognosis of MET-overexpressing non-small cell lung cancer (NSCLC) in a real-world patient cohort. Poster presented at: ESMO Congress 2023; October 20-24, 2023; Madrid, Spain. 11. Gherardi E, Birchmeier W, Birchmeier C, Vande Woude G. Targeting MET in cancer: rationale and progress. Nat Rev Cancer. 2012;12(2):89-103. 12. Referenced with permission from the NCCN Clinical Practice Guidelines in Oncology (NCCN Guidelines®) for Non-small Cell Lung Cancer V.2.2024. © National Comprehensive Cancer Network, Inc. 2024. All rights reserved. Accessed February 9, 2024. To view the most recent and complete version of the guideline, go online to NCCN.org. 13. Pawelczyk K, Piotrowska A, Ciesielska U, et al. Role of PD-L1 expression in non-small cell lung cancer and their prognostic significance according to clinicopathological factors and diagnostic markers. Int J Mol Sci. 2019;20(4):824. 14. Thai AA, Solomon BJ, Sequist LV, Gainor JF, Heist RS. Lung cancer. Lancet. 2021;398(10299):535-554. 15. Tsao MS, Yatabe Y. Old soldiers never die: is there still a role for immunohistochemistry in the era of next-generation sequencing panel testing? J Thorac Oncol. 2019;14(12):2035-2038. 16. Lee GD, Lee SE, Oh DY, et al. MET exon 14 skipping mutations in lung adenocarcinoma: clinicopathologic implications and prognostic values. J Thorac Oncol. 2017;12(8):1233-1246. 17. Shi M, Ma J, Feng M, et al. Novel MET exon 14 skipping analogs characterized in non-small cell lung cancer patients: a case study. Cancer Genetics. 2021;256-257:62-67.
